Kracken
Kracken is a piece of software that can be used to quickly analyse the organisms present in your sequencing run. Kracken breaks the reads up into kmers and matches these to a pregenerated database. The database is located at /opt/storage2/ernest/kraken/kraken/standard_db
Here is an example command:
kraken --db /opt/storage2/ernest/kraken/kraken/standard_db --threads 5 --fastq-input --gzip-compressed <prefix>_1.fastq.gz <prefix>_2.fastq.gz > <prefix>.kracken
cut -f2,3 <prefix>.kracken | ktImportTaxonomy - -o <prefix>.html
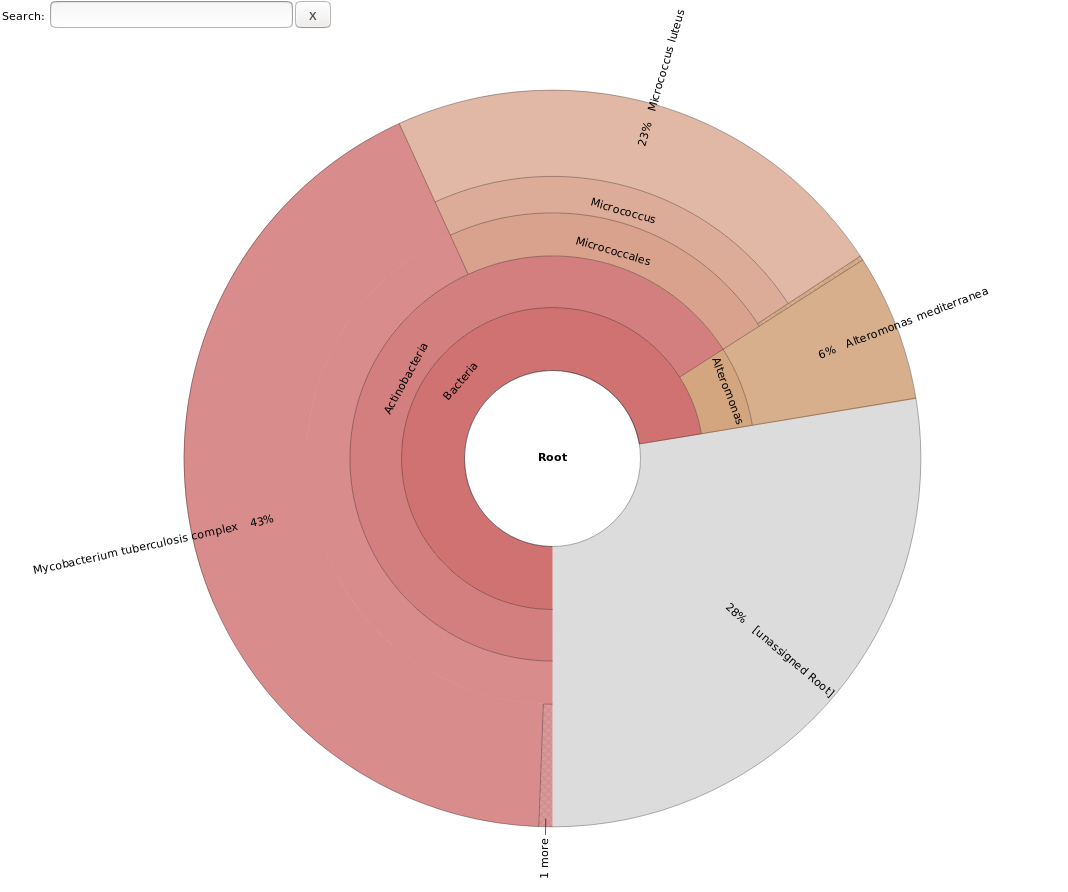
Where <prefix> should be replaced by whatever you have names your sample. Open the html file and check the results.
The results look like this: